With the advancement of research methods in biochemistry, the intracellular chemical processes have been gradually clarified. However, the traditional bulk biochemical experiments in test tubes may not reflect the complete processes in vivo owing to their inherent ensemble averaging. The test tube measurements provide averaged information obtained on large ensembles of molecules from many cells. Undoubtedly, the results deduced from the ensemble measurements are correct for depicting the behavior of the ensemble molecules, but on the other hand, the ensemble measurements hide the rare events and the variable properties of individual molecules. As opposed to test tube experiments, single-molecule techniques allow one to look beyond ensemble averages and thereby can reveal the events and properties that would otherwise be inaccessible, providing new insights for understanding the cellular physiological activities at individual cell/molecule levels. Consequently, utilizing single-molecule techniques to investigate the behavior of individual cells/molecules has become a new research hotspot in life sciences.
Atomic force microscopy(AFM) is a new single-cell single-molecule technique which was invented in 1986. Compared with traditional biochemical experimental methods, AFM has many advantages, such as it can work in fluids which is necessary for living cell experiments, it has nanometer resolution which makes us can observe the ultra-structures of the cell, and the sample preparation is easy. Because of these merits, AFM has attracted the attention of many researchers in the world to perform single-cell single-molecule experiments and these experiments have obtained impressive results, improving our understanding of cellular physiological activities. However, most of current AFM experiments were performed on cells cultured in vitro(cell line) which are very different from the cells in vivo. Hence investigating the behavior directly on patient cells is of important significance.
Researchers from Shenyang Institute of Automation (SIA), Chinese Academy of Sciences used AFM to investigate the molecular force on lymphoma patient cells. The pathological cells were prepared from the bone marrow of B-cell lymphoma patients whose bone marrow was intruded by lymphoma cells. Figure 1 shows the fluorescence staining images of cells from lymphoma patients. The optical images of the pathologic bone marrow sample from lymphoma are provided in Fig.1 A and D. The corresponding fluorescence images are provided in Fig.1 B and E. The cancer cells can be recognized by their special morphology. Then the drug molecules (antibodies) were linked onto the AFM tip surface to measure the interaction forces between target and drug. In order to verify whether antibodies had been linked onto the AFM tip surface, we performed SEM and fluorescence experiments, as shown in Fig. 2. From the SEM images, there were many particles on the surface of the functionalized tip while the surface of the normal tip was smooth. From the fluorescence images, the fluorescence of functionalized tip was bright, while the fluorescence of the normal tip was dim. The results of SEM and fluorescence experiments indicated that drug molecules were linked onto the surface of tip. Then the functionalized tip was used to measure the molecular binding force on patient cells, as shown in Fig.3. Fig.3A was a typical force curve where the abrupt peak indicated the target-drug binding. Fig.3B was a typical force curve where no target-drug binding occurred. The tiny peak of Fig.3B indicated the non-specific binding. Fig.3C was the histogram of the binding forces of one cell and Gaussian fit indicated that the binding force was 132±51pN. Fig.3D shows the binding forces of 22 different cells. The binding forces were mainly in the range of 110-140pN. The results improve our understanding of the behavior of patient cells and will facilitate further researches of antibody drug’s effect.

Figure 1 Fluorescence staining of cell sample from lymphoma patients. (A), (D) were optical images of the sample and (B), (E) were the corresponding fluorescence images, respectively. The insets in (A), (D) were the higher resolution optical images. (C), (F) were the higher resolution fluorescence images of the cancer cells indicated by the circle in (B), (E), respectively. (Image provided by LI Mi, et.al)
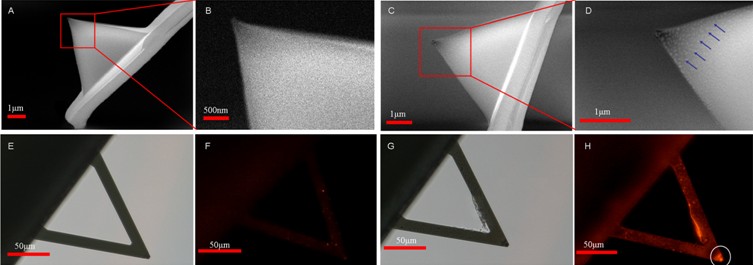
Figure 2 Using SEM and fluorescence microscopy to verify whether rituximabs had been linked onto the AFM tip. (A), (B) were the SEM images of the normal probe and (C), (D) were the SEM images of the functionalized probe. (B), (D) were the higher resolution SEM images (indicated by the squares in (A), (C), respectively). (E), (G) were the optical images of the normal probe and the functionalized probe respectively and (F), (H) were the corresponding fluorescence images. (Image provided by LI Mi, et.al)
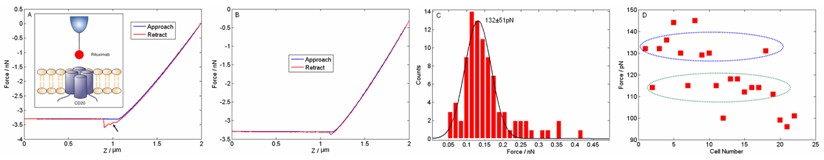
Figure 3 Measuring the drug-target binding forces on pathological cells. (A) A typical force curve with target-drug binding occurred. (inset) Principle of the force measurement. (B) A typical force curve with no binding occurred. (C) Histogram of binding forces of one cell. (D) The distribution of the binding forces of 22 different cells. (Image provided by LI Mi, et.al)
This work was published on Scanning, volume 35, 2013, 40-46. It was partly supported by the National Natural Science Foundation of China (60904095, 61175103).
CONTACT:
LI Mi
Shenyang Institute of Automation, Chinese Academy of Sciences
E-mail: limi@sia.cn